
Order the NovaSeq X Series
Advanced chemistry, optics, and informatics combine to deliver exceptional speed and data quality, outstanding throughput and scalability.
A high-performance, fast, and reliable human whole-exome sequencing solution that includes a comprehensive, up-to-date exome enrichment panel.
Assay time
Hands-on time
Input quantity
Illumina DNA Prep with Exome 2.5 Enrichment offers a cost-effective, reliable solution for human whole-exome sequencing (WES). This kit provides:
This whole-exome sequencing kit includes library prep and hybridization reagents, clean up/size selection beads, and indexes. The kit also includes the Twist Bioscience for Illumina Exome 2.5 Panel, which offers improved target region coverage for variants reported in public databases.
| Assay time | 6.5 hr |
|---|---|
| Automation details | Explore available automation methods |
| Content specifications |
37.5 Mb coding content (≥ 99% of RefSeq, CCDS, ClinVar and ACMG pathogenic/likely pathogenic variants, COSMIC Cancer Gene Census), OMIM |
| Description | A fast and easy-to-use library prep with enrichment workflow with a focused enrichment probe panel of up-to-date exome content for cost-effective and reliable human whole-exome sequencing. |
| Hands-on time | ~2 hr |
| Input quantity | 50-1000 ng high-quality genomic DNA (For direct blood and saliva, see reference guide) |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NextSeq 550Dx in Research Mode, NovaSeq X System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Bead-linked transposome and hybrid-capture chemistry |
| Method | Target Enrichment, Exome Sequencing, Targeted DNA Sequencing |
| Multiplexing | Pre-enrichment pooling of up to 12-plex is tested. Kit configuration supports multiplexing up to 192 samples. Up to 384 possible. |
| Nucleic acid type | DNA |
| On-target reads | ~ 90% based on average performance from internal testing |
| Specialized sample types | Blood, Saliva |
| Species category | Human |
| Technology | Sequencing |
| Uniformity | ≥ 91% (% coverage at 20× for 5 Gb) based on average performance from internal testing |
| Variant class | Single nucleotide polymorphisms (SNPs), Loss of heterozygosity (LOH), Somatic variants, Germline variants, Insertions-deletions (indels), Copy number variants (CNVs) |
Illumina DNA Prep with Exome 2.5 Enrichment kits include library prep, enrichment, enrichment probe panel, purification beads, and index adapter reagents. Choose a different index set if preferred.
The Twist Bioscience for Illumina Mitochondrial Panel is an optional fixed content spike-in panel that allows enrichment and sequencing of mitochondrial DNA variants.
For direct blood input, the Flex Lysis Reagent Kit is required.
The Infinium MIDI Heatblock Insert fits in the SciGene HybEx Microheating system and is required for the enrichment step.
The Illumina DNA Prep with Exome 2.5 Enrichment is a fast and comprehensive end-to-end whole-exome sequencing solution that enables support from sample to reporting.
Illumina DNA Prep with Exome 2.5 Enrichment
Illumina Connected Insights - Somatic
Not available in all countries. Illumina Connected Insights supports user-defined tertiary analysis through API calls to third-party knowledge sources.
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 2000 System | Samples per run (by flow cell type): P2: 19, P3: 57 (based on 50× mean target coverage, 42M reads, 4.2 Gb of data) |
2 × 101 bp |
| NovaSeq 6000 System | Samples per run by flow cell type: SP: 34, S1: 69, S2: 176, S4: 428 based on 50× mean target coverage, 42M reads, 4.2 Gb of data. Note: S4: 428 requires NovaSeq Xp workflow or additional indexes. |
2 × 101 bp |
Whole-exome sequencing is used to investigate protein-coding regions of the genome to uncover genetic influences on disease and population health.
Complex and genetic disease research
Comprehensive array and next-generation sequencing solutions to accelerate research of various genetic complex diseases.
Our sequencing and microarray technologies support a broad range of cancer genomics research applications, from DNA to RNA analysis, epigenetics, and more.
| Illumina DNA Prep with Exome 2.5 Enrichment | Illumina DNA Prep with Enrichment | |
|---|---|---|
| Assay time | 6.5 hr | ~6.5 hr |
| Automation details | Explore available automation methods | Explore available automation methods |
| Content specifications |
37.5 Mb coding content (≥ 99% of RefSeq, CCDS, ClinVar and ACMG pathogenic/likely pathogenic variants, COSMIC Cancer Gene Census), OMIM |
Exome: 45 Mb exonic content (≥98% of RefSeq, CCDS, and Ensembl coding content). Custom: 0.5 - 15 Mb genomic content of interest. Fixed panels: Content varies by panel. |
| Description | A fast and easy-to-use library prep with enrichment workflow with a focused enrichment probe panel of up-to-date exome content for cost-effective and reliable human whole-exome sequencing. |
Prepare sequencing libraries for a variety of experiments, including whole-exome, custom, and fixed panel targeted sequencing, from low DNA inputs and a variety of sample types in ~6.5 hours. |
| Hands-on time | ~2 hr | ~2 hr |
| Input quantity | 50-1000 ng high-quality genomic DNA (For direct blood and saliva, see reference guide) | 10-1000 ng high-quality genomic DNA or 50-1000 ng FFPE DNA. (For blood and saliva, see the reference guide). |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NextSeq 550Dx in Research Mode, NovaSeq X System, NovaSeq 6000 System, NovaSeq X Plus System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, MiSeqDx in Research Mode, MiniSeq System, NextSeq 550Dx in Research Mode, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Bead-linked transposome and hybrid-capture chemistry | Bead-bound transposomes and hybrid-capture chemistry |
| Method | Target Enrichment, Exome Sequencing, Targeted DNA Sequencing | Target Enrichment, Exome Sequencing, Custom Sequencing, Targeted DNA Sequencing |
| Multiplexing | Pre-enrichment pooling of up to 12-plex is tested. Kit configuration supports multiplexing up to 192 samples. Up to 384 possible. | Up to 384 available unique dual indexes. Pre-enrichment pooling of up to 12-plex is tested. |
| Nucleic acid type | DNA | DNA |
| On-target reads | ~ 90% based on average performance from internal testing | ≥85% for exome panel (results from example data set; actual performance may vary) |
| Specialized sample types | Blood, Saliva | Blood, Low-Input Samples, FFPE Tissue, Saliva |
| Species category | Human | Other, Human |
| Technology | Sequencing | Sequencing |
| Uniformity | ≥ 91% (% coverage at 20× for 5 Gb) based on average performance from internal testing | ≥90% (% coverage at 20x for 4 Gb; for exome panel) |
| Variant class | Single nucleotide polymorphisms (SNPs), Loss of heterozygosity (LOH), Somatic variants, Germline variants, Insertions-deletions (indels), Copy number variants (CNVs) | Single nucleotide polymorphisms (SNPs), Loss of heterozygosity (LOH), Somatic variants, Germline variants, Insertions-deletions (indels), Copy number variants (CNVs) |
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
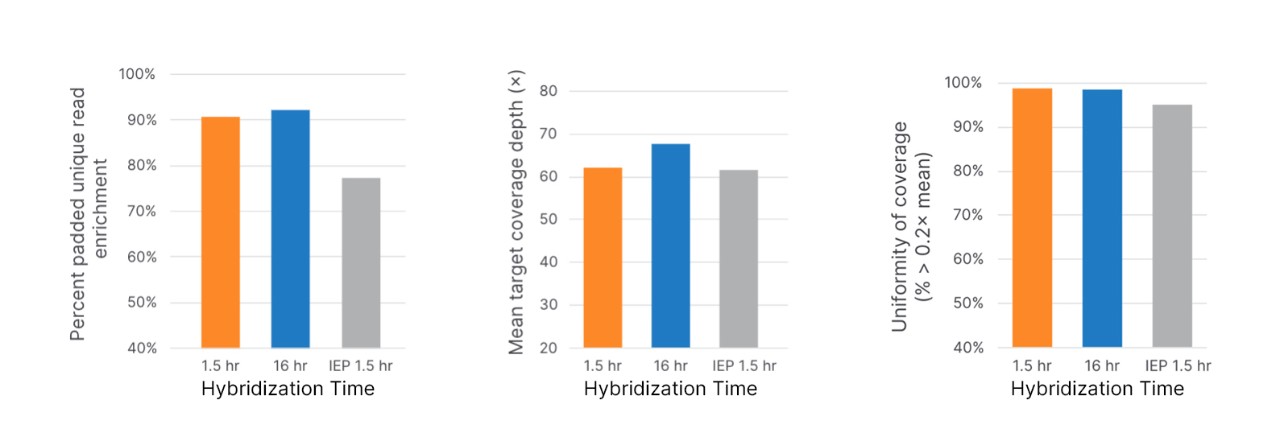

Libraries were constructed and enriched using Illumina DNA Prep with Exome 2.5 Enrichment with either the included Exome 2.5 Panel with a 1.5 hr or 16 hr hybridization time or the Illumina Exome Panel (IEP, 20020183) with a 1.5 hr hybridization time. Enriched libraries were sequenced on a NovaSeq 6000 System and analyzed using DRAGEN Enrichment App.
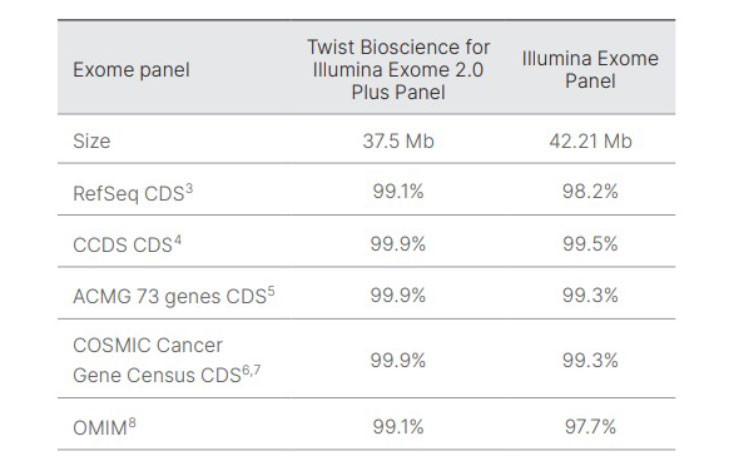

The bed files for the Exome 2.5 Panel and Exome Panel were compared to the listed public databases and the percent overlap was determined and reported.
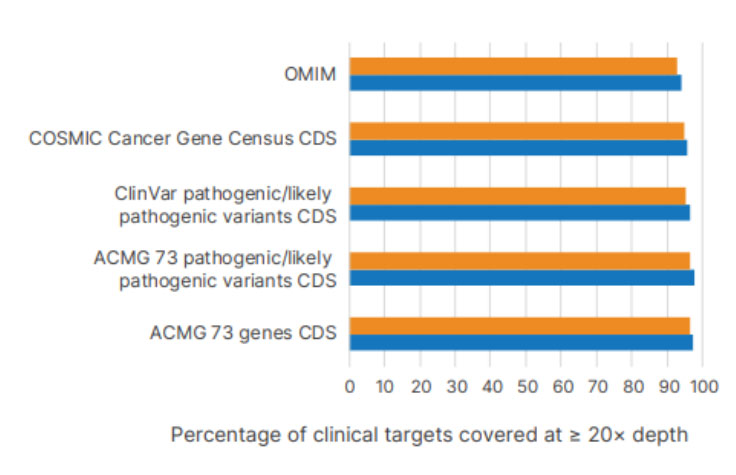

Illumina DNA Prep with Exome 2.5 Enrichment using a 1.5-hr (orange) or 16-hr (blue) hybridization time shows high average percent coverage at ≥ 20× of targets from public databases.
Both high- and low-throughput walk-away automation methods are available through our automation partners to reduce hands-on time and minimize errors.
Illumina DNA Prep with Exome 2.5 Enrichment, (S) Tagmentation Set B (96 Samples, 12-plex)
20077595
Includes Illumina DNA Prep with Enrichment library preparation and enrichment reagents, Twist Bioscience for Illumina Exome 2.0 Plus Panel, Illumina Purification Beads, and IDT for Illumina DNA/RNA UD Indexes Set B, Tagmentation for whole exome enrichment of 96 libraries (eight, 12-plex enrichment reactions).
List Price:
Discounts:
Illumina DNA Prep with Exome 2.5 Enrichment, (S) Tagmentation Set D (96 Samples, 12-plex)
20077596
Includes Illumina DNA Prep with Enrichment library preparation and enrichment reagents, Twist Bioscience for Illumina Exome 2.0 Plus Panel, Illumina Purification Beads, and IDT for Illumina DNA/RNA UD Indexes Set D, Tagmentation for whole exome enrichment of 96 libraries (eight, 12-plex enrichment reactions).
List Price:
Discounts:
Twist Bioscience for Illumina Mitochondrial Panel
20093180
Includes Twist Bioscience for Illumina Mitochondrial Panel, 32 ul of reagent for 96 samples (8 hybridization reactions with 12 samples per reaction). This panel can be used with the Exome 2.5 Panel in the Illumina DNA Prep with Exome 2.5 Enrichment kits, for combined enrichment of both human exome plus mitochondrial DNA from whole genome libraries. The Mitochondrial Panel is sold as an accessory for the Exome 2.5 Panel products.
List Price:
Discounts:
Flex Lysis Reagent Kit (96 reactions)
20018706
Includes reagents for processing blood samples with the Illumina DNA Prep and Illumina DNA Prep with Enrichment. Purchase library prep, enrichment, enrichment probe panel, and index adapter reagents separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set A, Tagmentation (96 Indexes, 96 Samples)
20091654
Illumina® DNA/RNA UD Indexes Set A, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set B, Tagmentation (96 Indexes, 96 Samples)
20091656
Illumina® DNA/RNA UD Indexes Set B, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set C, Tagmentation (96 Indexes, 96 Samples)
20091658
Illumina® DNA/RNA UD Indexes Set C, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set D, Tagmentation (96 Indexes, 96 Samples)
20091660
Illumina® DNA/RNA UD Indexes Set D, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Infinium MIDI Heatblock Insert
BD-60-601
Heatblock insert for the SciGene Hybex Microheating system required for the Nextera Flex for Enrichment hybridization steps.
List Price:
Discounts:
Illumina Purification Bead, 100mL
20060057
Includes one, 100-ml container of beads for library clean-up and size selection. Purchase library prep and enrichment reagents, probe panels, and indexes separately.
List Price:
Discounts:
Illumina Purification Bead, 400mL
20060058
Includes four, 100-ml container of beads for library clean-up and size selection. Purchase library prep and enrichment reagents, probe panels, and indexes separately.
List Price:
Discounts:
Showing of
Product
Qty
Unit Price
Product
Catalog ID
Quantity
Unit price
Yes, gDNA input of 10-49 ng can be used. However, this protocol does not normalize final pre-enrichment library yields < 50 ng and, therefore, quantification and normalization of libraries before and after enrichment is required.
This protocol has been validated with fresh blood collected in EDTA collection tubes. Other anticoagulants may be used but are not guaranteed when using this kit.
This protocol is validated for saliva collected only in Oragene DNA Saliva collection tubes.
The Twist Bioscience for Illumina Mitochondrial Panel is an optional fixed content panel designed to cover the 16,659 base pairs (bp) and 37 genes of the mitochondrial genome (chrM) and allows enrichment and sequencing of mitochondrial DNA (mtDNA) variants. Use this panel as a spike-in panel in the Illumina DNA Prep with Exome 2.5 Enrichment protocol and add coverage of ChrM to your experiments.
The products previously known as Illumina DNA Prep with Exome 2.0 Plus Enrichment are now called Illumina DNA Prep with Exome 2.5 Enrichment (Cat. No. 20077595 and 20077596). The exome panel oligos have not changed, but the included indexes have been updated to Illumina DNA/RNA UD Indexes.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.