How to interpret DNA strand and allele information for Infinium genotyping array data
09/03/21
When comparing genotyping data, it is important to use the same DNA strand designation. For Infinium array data analyzed using GenomeStudio, sample genotypes can be reported in several formats, including Plus Strand, Top Strand, and/or Forward Strand. The strand designations are based on information provided in the product manifest file, and are relative to the nucleotides reported in the SNP column.
Note: The first nucleotide listed for the SNP is Allele A, and the second nucleotide is Allele B. Alleles A and B are determined by the Top/Bottom, A/B designation (for more details, see the bulletin Simple Guidelines for Identifying Top/Bottom TOP/BOT Strand and A/B Allele and the Technical Note "TOP/BOT" Strand and "A/B" Allele).
DNA strand designations
- Top/Bottom (TOP/BOT, T/B): This designation was developed by Illumina to allow unambiguous specification of the strand, based on the variant and surrounding DNA sequence, without reference to any database (dbSNP, genomic reference, etc.). The T/B annotation within the IlmnID provides the strand designation of the SNP column in the manifest. It is useful when no reference is available, or when comparing to data generated prior to a reference being established.
- Plus/Minus (+/-): Plus strand corresponds to the genomic sequence as reported in the FASTA for the genomic reference. The +/- strand designation for the SNP column is provided in the RefStrand column of the manifest for commercial arrays, and for custom arrays by request. These designations can change with NCBI Genome Build versions.
- Forward/Reverse (FWD/REV, F/R): The Illumina designation of Forward strand indicates that the alleles in the SNP column match the RefSNP alleles displayed in dbSNP. The F/R annotation is found within the IlmnID. Neither this Illumina strand designation nor a Forward strand report from GenomeStudio are expected to correspond to the FWD/REV strand designation in dbSNP, which provides the orientation of the RefSNP alleles relative to the genomic reference. For custom content, the customer defines this annotation.
GenomeStudio final report strand options
- Allele1, 2 – Top, Plus, and/or Forward: Allele 1 and 2 nucleotides correspond to Allele A and B, and are reported on the Top, Plus, or Forward strand as described above.
- Allele1, 2 – Design: Allele 1 and 2 nucleotides correspond to Allele A and B, and are reported on the strand on which the probe was designed. This is generally not a useful convention for sharing data; however, may be useful for investigation of data in which multiple designs have been created for the same variant.
- ILMN Strand: The strand used to design each probe is designated as TOP/BOT for SNPs or PLUS/MINUS for indels as indicated in the ‘IlmnStrand’ column of the manifest. The alleles in the ‘SNP’ column are on the IlmnStrand.
- Customer Strand: The strand submitted to the Illumina designer by Illumina or a customer is designated as TOP/BOT for SNPs or PLUS/MINUS for insertion/deletion (indel), as indicated in the ‘SourceStrand’ column of the manifest.
- Plus/Minus Strand: The +/- strand designation for the SNP column is indicated if provided in the ‘RefStrand’ column of the manifest.
Example strand annotations for rs10000030
- Illumina Manifest:

- dbSNP database:


The rs10000030 strand designations for the [T/C] SNP reported in the SNP column can be determined by referencing the IlmnStrand and RefStrand columns in the manifest.
- [T/C] strand = BOT (IlmnID and IlmnStrand), Minus (RefStrand), and R (IlmnID, SNP is on the opposite strand of the dbSNP entry)
- Therefore, the [A/G] strand = TOP, Plus, F (SNP matches dbSNP entry)
- The SNPs are listed in A/B allele order according to the TOP/BOT convention, rather than the Ref/Alt allele order.
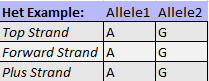
In summary, the following relationships hold true for dbSNP variants within the Illumina manifest, except in cases of unmapped probes.




